PUMAdb : Help Entering Results for a Batch of Arrays
Your session is inactive. Login
Contents
Many times, you will have enough microarray results to enter that entering the file names one-by-one on a web page will be too time consuming. If this is the case, you can construct a file that provides all the information you would otherwise enter on the web page. The Data Entry for Microarray Experiment form is used to enter a batch of experiments into the database according to instructions specifed in the batch file. This help describes the information you need to enter an experiment and how to create the batch file. Small numbers of experiments can also be entered individually, and a separate help is available for that procedure.
To load a group of experiments into the database, you first need to assemble a batch file in which each line contains all the information needed to enter one experiment. This is the same information that goes on the web form for individual experiment entry. The batch file must be tab delimited and have the lists below in the header. These lists are the same as the entry box titles on the individual experiment entry form. The columns in the batch file can be in any order.
Within the File menu of your browser, Just "Save As Text" and either copy or edit the resulting file. Do not change the name or spelling of the labels. All fields are required except those marked optional. Optional fields may be ommitted entirely from the batch file, or contained with empty values. Your completed batch file must be in your loader account before you can enter the data. Data files must also be in your loader account. All fields marked "optional" may be left blank or not included in your batch file. All fields not marked as optional are required.
| AFFYMETRIX batch file columns (sample) | AGILENT batch file columns (sample) |
|---|---|
|
|
| GENEPIX / SCANALYZE / SPOTREADER batch file columns (sample) | NIMBLEGEN batch file columns (sample) |
|
|
1Add to Expt is used if you are adding a new result set to an existing experiment. The value must be "Y". The values for Experiment Name and Slide Name must be the same as the experiment to which you are adding the result set.
2Data files can be identified in the batch file by filenames only if the batch file is in the same directory as all the data files. If some data files are organized in subdirectories inside incoming/, then the batch file should include the path to those data files relative to the batch file. If, eg. some of the data files are in the "worm_aging" directory inside the incoming/ directory, the path would be: "worm_aging/1234.gpr".
3Experiment Date will default to the date the experiment is entered if the column is left blank. Two date formats are accepted. One is a 4-digit year, 2-digit month, and 2-digit day (YYYY-MM-DD, e.g. 2006-11-22), and the other is the Excel default (MM/DD/YY e.g.11/22/06).
4Normalization Type is required and can be "Computed", "Regression" or "User Defined". If Normalization Type is "User Defined", then Norm Value is required. Any number can be entered as a Norm Value. If norm Type is "Computed," then the default computed normalization is used. If the Normalization Type is either "Computed" or "Regression", the Norm Value column should be left blank. Normalization type is required for entering Agilent and Affymetrix data, but is ignored.
5Collaborative Group and Individual User are groups and/or users to whom you give permission to view this experiement. They must exist in the User Groups and the Users list, respectively. Separate multiple values with commas.
6Green Scan File and Red Scan File (Agilent Only): if you have used TiffSplitter to split your image into two files, then enter each name name in these files. If you have NOT split the image, then enter its name in the Green Scan File and leave the Red Scan File blank. A single batch file must contain ALL split images or ALL unsplit images, not a combination of the two.
Example batch files from which you can copy headings are available for:
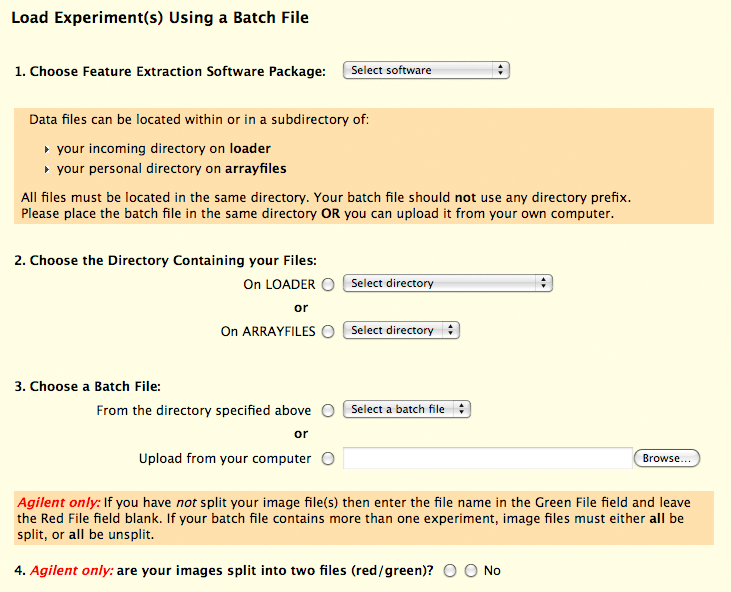
Initiating the Process: Choosing "Load Experiment(s) using a Batch File" from the Experiment and Result Entry form takes you to the batch entry form. Here you specify the feature extraction software, choose the directory containing your data files and select the batch file. (Alternatively, you can upload the batch file from your own computer.) For convenience, files containing the word "batch" are placed at the top of the selection list and so you might want to include the word "batch" in the filename, i.e., "affy_batch.txt". (The filename can have any extension.) Now you can choose to:
Experiment loading is commenced by entering your loading-data into a
queue. The rate of loading is determined by a number of factors,
including both the load on the database and how many other array-load
requests were made prior to yours. If there are no delays, it usually
takes at least five minutes per array, but can take
quite a bit longer if your arrays have a large number of spots (human arrays) or
if many other users are using the database. An Affymetrix tiling array can
take up to one hour to load into the database and so as not to slow the
database to a crawl, only one tiling array can be run at once; all other jobs are queued.
During this time, you can check the progress of your experiment load
within the queue. After your data is successfully entered into the
queue (note: this is not the same thing as final entry into the
database), you should receive a confirmation screen as well as an
email notifying you:
Your database entry request (batch number XXXX) has been queued for loading. Please note the data for your array(s) ARE NOT YET IN THE DATABASE. Do NOT delete any of your files until you receive email confirmation that the data have been loaded. Progress of is batch within the queue can be viewed at: http://puma.princeton.edu/cgi-bin/tools/queue/nph-ProgressQuery?batchno=XXXX If you have any questions please contact the database curators array@princeton.edu)
You can check the progress of your experiment load based on the batch number reported to you with either the link on the queue confirmation page or from the URL in the email.
If all goes well, you will eventually get an email message that says:
Loading of your array data (batch number XXXX) has completed.
1 out of 1 were successfully loaded.
Details of the load process have been written to:
/loader/ftphome/username/logs/XXXX.log,
or you can temporarily view the details via the web at:
http://puma.princeton.edu/cgi-bin/tools/queue/nph-ProgressQuery?batchno=XXXX
If you have any questions please contact the curators
(array@princeton.edu).
At the bottom of the HTML confirmation page or in the log file in your logs directory on loader.princeton.edu should be the message:
==== 1 out of 1 were successfully loaded. ====