Contents
PUMAdb generates .pcl files when data are retrieved from the database. After clustering a .cdt file is generated, and in addition, .gtr and .atr files may also be generated. The complete dataset without any processing can also be downloaded as an Excel file.
The complete normalized dataset without any additional filtering can be downloaded as an .xls file, that is directly readabel by Excel. To download this file, select the experiment on the Basic or Advanced Search page and select the display data button. By selecting the 'Download Raw Data' icon (  ) from the table you can download the Excel file.
) from the table you can download the Excel file.
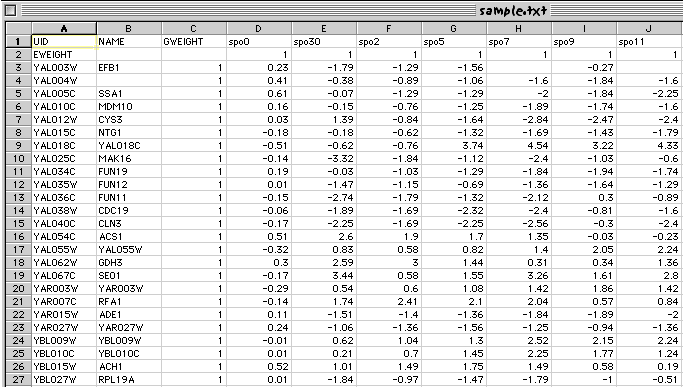
The pcl file is a tab-delimited pre-clustering file. The first three columns are as follows:
This column will contain a name ascribed to the entity on that row, such as ORF name, or CLONEID. The column itself can be named anything, but by convention is named YORF when it contains yeast ORF names, CLID when it contains cloneids, and LUID when it contains LUIDs. These name are simply so that after clustering, the Treeview software can use the contents of the column in URLs without additional configuration. This column MUST contain some text on every row.
This column can contain descriptive information about the entity, eg process or function, or gene symbol. It too can be named anything. It can optionally be left blank, but the column itself must be present.
This column allows you to weight genes with different weights, for instance if a gene appears on an array twice, you may want to give them a weight of 0.5 each. For the most part people leave this column with a value of 1 for every gene. This column must be present, and each row must have an entry.
In addition the file must begin with the following two rows:
This contains the column headers as described above for columns 1, 2 and 3, then contains the experiment names for all the data columns that exist in the file. Each data column must have a text entry as a name for that column.
This is the EWEIGHT row. The entry in the first column for this row should say EWEIGHT, then for each experiment, there should be an EWEIGHT value. This will usually be 1, but if the same experiment is duplicated twice, you may want to give these repeats an EWEIGHT of 0.5.
The remaining cells in the file contain the actual data, such that the row and column specifies to which gene and which experiment a particular piece of data corresponds. If you had modified, or created your pcl file in Excel, it would look something like this:
You should then choose Save As... from the File menu, and elect to the the file as type Text (Tab delimited), as indicated below:

In general the pcl file will contain log-transformed data, which is needed for clustering to work properly.
Extended pcl splits concatenated annotations from the "Name" column into individual columns. The "Name" column is replaced with
the name of the first annotation. Below, in the regular pcl format, the "Name" column contains the gene name, coordinates, and the
number of rows collapsed by systematic name.

In the extended pcl, the "Name" column is replaced by "Gene_Name"; "Map_Coords" and "# Collapsed" are put in new columns.

At this time, extended pcl files cannot be loaded into the PUMA repository.
When you cluster a .pcl file you will generate a .cdt (clustered data table) file, which will contain the original data, but reordered, to reflect the clustering. In addition, if you clustered by genes, you will get a .gtr file (gene tree), and if you clustered by experiments you will get a .atr file (array tree). These tree file reflect the history of how the cluster was built, and can be used to contruct how the tree(s) should look.
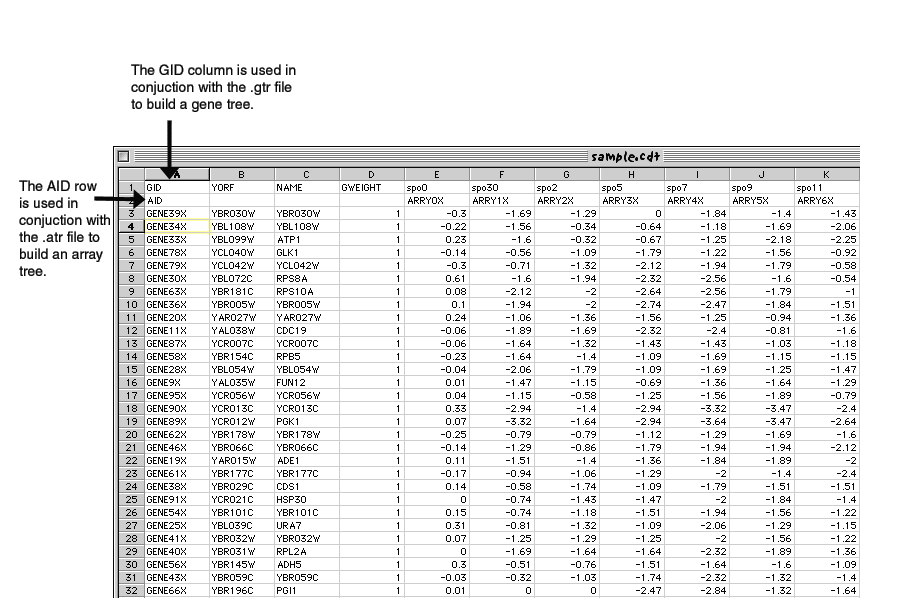
The .gtr (gene tree) file records the order in which the genes (rows) were joined during clustering.

The .atr (array tree) file records the order in which the arrays (columns) were joined during clustering.

After the printlist (aka godlist) has been entered into PUMAdb, it is available for download in a tab delimited text file format with the extension: .gdl. This file contains all the availble information about the particular print. Select the appropriate print from the following page: Print List.
A subset of the data contained in the godlist file can be downloaded in a format the can be uploaded in GenePix. These .gal (GenePix Array List) files describe the names and identifiers of the printed substances associated with each spot of a given sector, row and column. You can download the .gal file by selecting the appropriate print from the following page: Print List. The GAL format is defined here.